- Research
- Open access
- Published:
Using meta-analysis and CNN-NLP to review and classify the medical literature for normal tissue complication probability in head and neck cancer
Radiation Oncology volume 19, Article number: 5 (2024)
Abstract
Purpose
The study aims to enhance the efficiency and accuracy of literature reviews on normal tissue complication probability (NTCP) in head and neck cancer patients using radiation therapy. It employs meta-analysis (MA) and natural language processing (NLP).
Material and methods
The study consists of two parts. First, it employs MA to assess NTCP models for xerostomia, dysphagia, and mucositis after radiation therapy, using Python 3.10.5 for statistical analysis. Second, it integrates NLP with convolutional neural networks (CNN) to optimize literature search, reducing 3256 articles to 12. CNN settings include a batch size of 50, 50–200 epoch range and a 0.001 learning rate.
Results
The study's CNN-NLP model achieved a notable accuracy of 0.94 after 200 epochs with Adamax optimization. MA showed an AUC of 0.67 for early-effect xerostomia and 0.74 for late-effect, indicating moderate to high predictive accuracy but with high variability across studies. Initial CNN accuracy of 66.70% improved to 94.87% post-tuning by optimizer and hyperparameters.
Conclusion
The study successfully merges MA and NLP, confirming high predictive accuracy for specific model-feature combinations. It introduces a time-based metric, words per minute (WPM), for efficiency and highlights the utility of MA and NLP in clinical research.
Introduction
Advancements in radiation therapy techniques for head and neck cancer have significantly improved patients' quality of life [1]. However, potential complications such as dysphagia, xerostomia, and mucositis can hinder recovery and amplify adverse effects. Specifically, radiation-induced xerostomia substantially diminishes patients' well-being, leading to oral health issues and communication barriers [2].
To enhance the welfare of head and neck cancer patients, researchers are exploring innovative approaches, including artificial intelligence (AI) and predictive algorithms, to investigate potential risk factors for complications. This multidisciplinary research has proliferated a vast body of publications. For instance, a literature search using the terms "artificial intelligence" and "head and neck cancer" between 2013 and May 2022 yielded 734,207 related articles on WOS, indicating a marked upward trend.
Given the sheer volume of published literature, comprehensive understanding through traditional literature reviews becomes increasingly challenging. Therefore, systematic search and filtering methods are crucial. Optimized strategies involve meta-analysis (MA) for synthesizing literature information, quantitatively integrating high-quality data to create valuable annotated datasets, thereby providing robust quantitative evidence for clinical decision-making.
However, conducting an integrated MA is time-consuming and labor-intensive, particularly in literature screening [3]. Reviewers face the daunting task of sifting through a plethora of articles with varying degrees of expertise and clinical relevance. To enhance the efficiency and accuracy of MA, this study employs natural language processing (NLP) techniques. As a significant branch of Artificial Intelligence, NLP enables computers to understand human language and has proven its applicability across various domains [4]. Utilizing NLP can augment the quantitative capabilities of MA, minimize human errors, and automate the screening process. The primary aim of this approach is to improve analytical efficiency while reducing human error.
NLP accelerates literature reviews by adeptly categorizing pertinent articles. Numerous studies have improved machine learning methods using publicly accessible literature from 15 systematic reviews [5,6,7,8]. For instance, Yujia et al. employed various machine learning models to classify abstracts into two categories related to cancer risk in genetic mutation carriers (penetrance) or the prevalence of genetic mutations [3]. Impressively, they achieved over 88% accuracy in both models. Zhengyi et al. demonstrated that NLP-based methods could substantially reduce the review workload while maintaining the ability to identify relevant research [3]. However, to date, no NLP techniques have been specifically tailored for literature on complications following head and neck cancer radiation therapy or normal tissue complication probability (NTCP). Furthermore, there's a conspicuous lack of an annotated dataset for crafting a machine learning model dedicated to discerning relevant articles in this domain.
Our research aims to fill this gap by creating an annotated abstract dataset focusing on the likelihood of three common complications post-radiation therapy for head and neck cancer—mucositis, xerostomia, and dysphagia. We will employ machine learning-based NLP methods to classify abstracts into this annotated dataset. The ultimate goal is to minimize human error and enhance analytical efficiency.
Materials and methods
Research framework
Our research process, based on MA, is divided into two parts, as depicted in Fig. 1. The first part employs MA to investigate NTCP predictive models for three common complications post-radiation therapy in head and neck cancer patients—xerostomia, dysphagia, and mucositis. The study encompasses patient demographics, methodologies, and outcomes, hypothesizing that significant variations may arise from different complication types, model choices, and predictive factors. By comparing various models and feature combinations, we aim to identify those with superior predictive capabilities, offering more effective predicting methods for clinical use. Statistical analyses are conducted using Python 3.10.5, with the null hypothesis stating that all model-feature combinations perform equally well in predicting complications, and the alternative hypothesis positing that at least one combination significantly outperforms the others.
The second part integrates natural language processing with convolutional neural networks (CNN) to enhance literature retrieval efficiency and result reliability. This approach aims to accelerate the time required for research on the NTCP of complications in head and neck cancer, offering quicker and more reliable insights for future studies and clinical applications.
Eligibility criteria, information sources, and search strategy
This study outlines the research content on head and neck cancer patients using the PICOS framework [9] (patient characteristics, intervention measures, control group, outcome), as showed in Fig. 2. Patient characteristics focus on head and neck cancer patients; interventions encompass all radiation therapy techniques for treating this cancer; control groups are categorized into machine learning, deep learning model types, and feature factors; and the outcome metric targets the AUC of multivariate NTCP models. Given its non-RCT or CCT nature, the study falls under the category of prospective trials.
After formulating the research theme, database searches are conducted using relevant keywords, covering both titles and abstracts. Primary search keywords are organized into three layers: patient, method, and outcome, and are explored in conjunction with the PICOS framework. To ensure completeness, Boolean "AND" searches are specifically performed for combinations of complications with AI and NTCP. Beyond the PICOS framework, the study also employs PubMed's MeSH terms and related literature to broaden its scope. Boolean logic and faceted search techniques are used to break down the indexing problem into multiple thematic layers, establish inter-layer relationships, and employ Boolean "OR" for union operations, ensuring the comprehensiveness of the search results (detailed keywords are provided in Additional file 1: Table S1) [10,11,12].
Selection process
Data extracted from each included study is determined through collaborative discussions among reviewers. One reviewer is responsible for data collection, while another performs cross-validation. The data encompasses authorship, publication year, types of complications, radiation therapy methods, employed models, features (prognostic factors), performance evaluation, as well as the study's contributions and conclusions.
Data extraction and risk of bias (RoB) assessment
In our study, when evaluating the quality and potential biases of the literature for MA, we opted for the PROBAST tool (Prediction model Risk Of Bias ASsessment Tool) over the commonly used Cochrane risk of bias assessment (RoB) tool. This strategic choice was influenced by the realization that a significant portion of the studies-included did not align well with the criteria of the Cochrane tool due to their unique characteristics.
PROBAST evaluates four domains: participants, predictors, outcome, and analysis. The participants domain assesses the representativeness of the target population and selection bias; the predictors domain evaluates the selection, relevance, reliability, and handling of predictive factors; the outcome domain focuses on the measurement and definition of outcomes, assessing their accuracy and consistency; and the analysis domain reviews methods for model development and validation, including sample size, missing data handling, model calibration, and discriminative ability.
Bias risk assessment is conducted using the PROBAST Excel interface developed by Borja M. Fernandez-Felix [13], with risk determinations—low, high, or unclear—derived from responses to signaling questions. An overall low risk is assigned only if all domains are low-risk; a single high-risk domain results in an overall high risk; and an unclear risk in one domain with low risk in others leads to an overall unclear risk. If all model domains are low-risk but lack external validation, the risk is elevated to high; however, if based on extensive data with internal validation, it can be considered overall low-risk.
Statistical methods
The MA in this study primarily contains the following key components:
-
I.
Study Selection and Features: Provides an overview of the included sample size, time frame, model characteristics, and predictive factors used.
-
II.
Combined Effect Size Results: Calculates the aggregated AUC, confidence intervals, and ANOVA p-values for the included studies, visually represented through forest plots to facilitate understanding of MA conclusions and statistical significance.
-
III.
Heterogeneity Test Results: Utilizes Cochran's Q statistic [14] and I2 values [15] to assess study heterogeneity. A low p-value in the Q statistic indicates the presence of heterogeneity, while a higher I2 value quantifies greater inter-study variability.
-
IV.
ANOVA analysis under random effects model: Calculates the effect size and variance for each study, using AUC as the benchmark. Determines the weight for each study, which is the reciprocal of the variance. Computes the overall effect size and variance. Calculates Q statistic, degrees of freedom, and I2 statistic. Conducts ANOVA analysis; if the Q statistic exceeds the degrees of freedom, significant inter-study differences exist, and F-values and p-values are calculated to assess the null hypothesis.
-
V.
This process covers study selection, effect size aggregation, heterogeneity testing, and variance analysis under a random effects model, offering a comprehensive evaluation of the predictive models' ability to forecast the incidence of complications.
Natural language processing (NLP) program design
To expedite the identification and retrieval of relevant literature while ensuring result reliability and accuracy, this study adopts a CNN for NLP, drawing inspiration from Yujia Bao's MA NLP model design [3]. This choice not only considers the nature of the data but also facilitates platform development, paving the way for the future integration of more deep learning models to enhance the classifier's accuracy and generalizability. In terms of abstract identification, the CNN model employed is capable of automatically learning language features from extensive text and achieving results across various tasks. Through word vector transformation and feature extraction, the CNN model effectively performs text classification and sentiment analysis. Key parameters used in this study include a batch size of 50, epoch range of 50–200, and a learning rate of 0.001.
Data preprocessing
The data preprocessing in this study is divided into two main phases. First, abstracts and titles that have undergone manual retrieval and initial screening are allocated into training, validation, and test sets. The positive and negative samples in the training and validation sets are distributed at a 2:8 ratio, while the test set is further fine-tuned to a more realistic 15:85 ratio to better reflect the prevalence of irrelevant samples. Second, for word vector embedding, the text is converted into jsonl format and manually annotated and cleaned, including the removal of potentially misleading punctuation and special characters. These preprocessing steps optimize the text for word vector embedding input in the CNN model, facilitating subsequent NLP and analysis.
Results
Literature review and research selection
After searching the WOS and PubMed databases, this study initially identified 3,256 potentially relevant articles, as illustrated in Fig. 3. The first round of screening, based on titles, eliminated studies unrelated to head and neck cancer or radiation therapy, leaving 87 articles for the second round. The second round, focused on abstracts, further excluded studies not involving head and neck or squamous cell cancer patients, or those not utilizing machine learning or deep learning as evaluation tools, resulting in 36 articles for full-text review. During this phase, articles not addressing predictions, not focusing on complications, or lacking AUC-related outcomes for multivariate NTCP models were also excluded, along with duplicates. Ultimately, 12 articles were included for review [16,17,18,19,20,21,22,23,24,25,26,27].
Performance of the CNN-NLP model
After comparing nine different optimizers, our study opted for Adamax (see Additional file 1: Table S2). With 50 epochs, Adamax achieved a Loss value of 0.51, an accuracy of 0.85, and an F1-Score of 0.75, along with a precision of 0.71. When the epochs were increased to 100, the accuracy and F1-Score improved to 0.87 and 0.79, respectively, while the precision reached 0.84. At 200 epochs, both accuracy and F1-Score peaked at approximately 0.94, clearly demonstrating the superior performance of the Adamax optimizer in the model.
After optimizer fine-tuning, as shown in Table 1, we evaluated coverage performance, which measures the overlap of identified studies under specific search subset conditions and assesses the efficacy of automated processing. We conducted tests on four different subsets, from WOS T1 to Pubmed T4, and compared the coverage rates when using Adam and Adamax optimizers across training cycles of 200, 100, and 50 epochs. In WOS T1, coverage was generally 0/9 regardless of the optimizer or training cycle, with Adam reaching a peak of 1/7 and low recognition frequency. In Pubmed T2, coverage was mostly 0/7, but a few articles were identified at epochs 100 and 50, not exceeding two in total. In WOS T3, Adam achieved a 3/4 coverage rate at 50 epochs, similar to Adamax. For Pubmed T4, Adam reached a 3/4 coverage rate at 100 epochs, while Adamax showed more stable performance across all training cycles, peaking at 2/4.
In the aspect of words per minute (wpm) for literature review, our study introduces a more objective method for time quantification. Beyond providing a standardized metric for future research, we also employ unit conversion and a deep learning-based Natural Language Text Classifier for temporal comparisons. In Table 2, we also calculated and compared the time spent on alternative tasks, converting wpm results to seconds, the details for the screening speed measured in WPM can be seen in Additional file 1: Table S3. We then contrasted this with the average time needed for text recognition during preprocessing in T1-T4 test sets using an Adamax-optimized CNN-NLP model. As shown in Table 1, despite considerations like text recognition capabilities, the time efficiency gained through NLP shows a significant, intuitive difference. (Code for WPM Calculation Algorithm captured from the monitor is shown in Additional file 1: Figure S1).
Features and model methods: systematic review
As shown in Table 3, the "studies-included" feature table aligns with the three dimensions of the MA issue discussed in our Materials and Methods section. In addition to the authors and publication years, the table also encompasses demographic characteristics, complications, types of radiation therapy techniques, algorithmic combinations in predictive models, predictive performance, and selected predictive factors. The systematic review ultimately included a total of 12 studies [16,17,18,19,20,21,22,23,24,25,26,27].
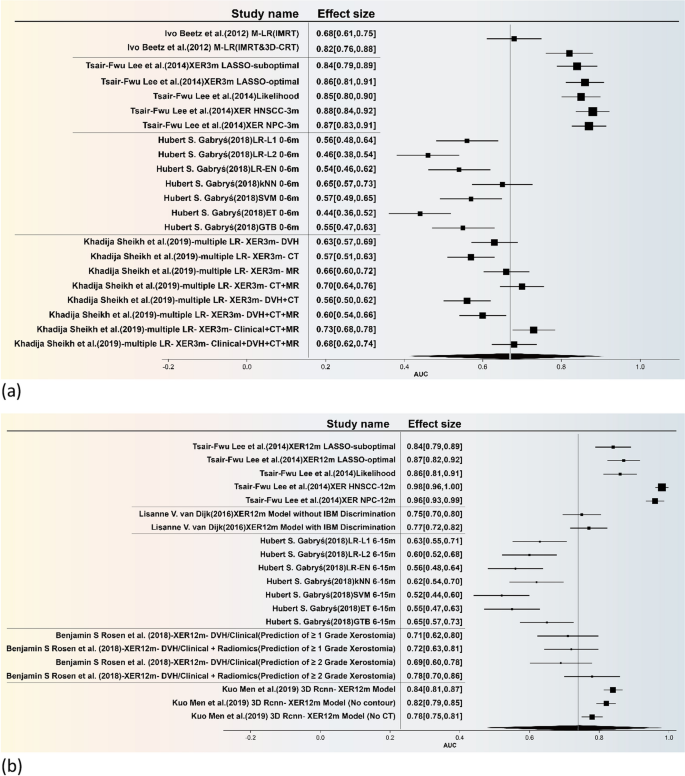
The forest plot is illustrated in Fig. 4, the present study undertakes a comprehensive and rigorous meta-analysis, focusing specifically on predictive models for xerostomia. Utilizing a feature table, we meticulously integrated the models employed across various studies and further stratified them into early and late phases for sub-group analysis. The combined effect sizes for these sub-groups are visually represented through forest plots (The funnel plot is included in Additional file 1: Figure S2). The temporal demarcation for these phases was set at six months, based on the seminal work of Hubert S. Gabryś [16].
Statistically speaking, the overall effect size for the Area Under the Curve (AUC) of early-effect xerostomia models (Fig. 4a) was 0.67, with a 95% Confidence Interval (CI) ranging from 0.40 to 0.91. This indicates that these models possess moderate predictive accuracy for early-effect xerostomia. However, the high heterogeneity, as evidenced by an I2 value of 80.32% and a Q-statistic of 5.34, suggests significant variability across different studies. For late-effect xerostomia (Fig. 4b), the overall AUC effect size was 0.74, with a 95% CI of 0.46 to 0.98. This result further corroborates the models' relatively high predictive efficacy for late-effect xerostomia. Nevertheless, the exceedingly high heterogeneity (I2 = 97.99%, Q-statistic = 52.48) implies that the applicability of these models may be limited across different research settings or patient populations.
In Table 4, titled "Prediction model Risk of Bias in Included Studies," the output for each question represents distinct focal points of work, encompassing a comprehensive evaluation of all critical stages in the development and application of prediction models as assessed by PROBAST. The assessment content is divided into four domains: 1. Participants, 2. Predictive Factors, 3. Outcomes, and 4. Analysis. These domains are further categorized based on three assessment outcomes, primarily labeled as "High Risk," "Low Risk," and "Unclear or Ambiguous."
Although the overall assessment reveals that only four studies exhibited low risk of bias in their data, with the remainder falling under high risk or unclear categories, it is noteworthy that in terms of applicability, only two included studies were assessed as having a higher risk, while two were categorized as unclear or ambiguous. This suggests that while there may be a pervasive issue of data bias, the applicability of these studies is less frequently compromised, thereby indicating a need for more rigorous methodological scrutiny to enhance the reliability and utility of future prediction models.
Discussion
Results of the MA study
In our study, we conducted a comprehensive retrospective analysis to evaluate AI-based predictive models for forecasting post-radiation complications like xerostomia in head and neck cancer patients. Our data revealed significant effect sizes of 0.67 and 0.74 for early and late-stage xerostomia, respectively, with p-values below 0.05, highlighting the distinctiveness of AI-based models in this context.
Interestingly, our findings contrast with earlier research by our team (Lee et al. [17, 18]) and Van Dijk et al. [19] We observed that incorporating image biomarkers, such as pre-processed CT data, did not necessarily enhance predictive accuracy compared to models solely based on traditional clinical factors and machine learning algorithms. This discrepancy may stem from variations in dataset composition and algorithmic parameters during model training and validation.
Further, research by Gabry et al. [16] identified key features like dosimetric shapes and salivary gland volume through algorithmic comparisons, reiterating the significant divergence between AI-based and traditional clinical models in xerostomia prediction.
However, our study also revealed certain limitations and challenges. Firstly, the limited scope of databases for literature search led to incomplete data and insufficient literature, restricting our ability to perform comprehensive meta-analyses and forest plot illustrations. Secondly, some studies lacked complete data, such as predictive confidence intervals, which further impacted our analysis. Just as per any other site, CNS NTCP literature suffers the same limitations, and no AI has been successfully implemented as yet [28]. Overall, while our study made progress in predicting normal tissue complications after radiotherapy for head and neck cancer, further research and validation are needed. Our findings align with Chulmin Bang's 2023 literature review, emphasizing that the clinical application of AI models still requires more in-depth exploration and validation [29].
Performance of the CNN-NLP model, optimizer optimization, and coverage
In this study, we presented an analysis focusing on the coverage rate of imbalanced datasets. Despite optimizing the algorithmic parameters, we abstained from employing data augmentation techniques like oversampling or undersampling to bolster the model's predictive accuracy. Our text classification model was conceptualized based on the research framework proposed by Yujia Bao, MA [3]. It's worth noting that this CNN-based model predominantly relies on abstracts rather than full texts for analysis. Consequently, the conversion rate of the included literature could be susceptible to variations in research themes and inclusion criteria, a limitation also acknowledged in Yujia Bao's work [3]. Nevertheless, recent advancements in large-scale language models such as GPT-3 and GPT-4 have shown capabilities in recognizing diverse file formats, including PDFs [30], and have exhibited remarkable precision in medical text identification [30, 31]. Progress has also been made in the realm of deep learning for medical text analysis, exemplified by CNN-based medical report retrieval studies [32]. These technological strides open new avenues for medical text identification, potentially mitigating the aforementioned limitations. We are currently exploring the development of models designed for automated full-text reviews to further enhance the comprehensiveness and accuracy of literature analyses.
Conclusion
In this study, we employ an integrative approach combining MA and NLP to explore feature factors for NTCP in head and neck cancer. Our results reject the null hypothesis \({H}_{0}\), confirming that specific model-feature combinations yield high predictive accuracy for identical complications. Utilizing CNNs in NLP, we streamline the meta-analytical process and introduce a time-based metric, words per minute (WPM) [33], for efficiency evaluation. This study underscores the utility of meta-analysis and NLP in clinical research, offering a methodological advancement for future studies aiming to optimize predictive models and operational efficiency.
Availability of data and materials
Not applicable.
References
Chen AM, et al. Quality of life among long-term survivors of head and neck cancer treated by intensity-modulated radiotherapy. JAMA Otolaryngol Head Neck Surg. 2014;140(2):129–33.
Gueiros LA, Soares MSM, Leao JC. Impact of ageing and drug consumption on oral health. Gerodontology. 2009;26(4):297–301.
Deng Z, et al. Validation of a semiautomated natural language processing–based procedure for meta-analysis of cancer susceptibility gene penetrance. JCO Clinic Cancer Inform. 2019;3:1–9.
Takeshita M, Rzepka R, Araki K. Speciesist language and nonhuman animal bias in English masked language models. Inf Process Manag. 2022;59(5):103050.
Jonnalagadda S, Petitti D. A new iterative method to reduce workload in systematic review process. Int J Comput Biol Drug Des. 2013;6(1–2):5–17.
Matwin S, et al. A new algorithm for reducing the workload of experts in performing systematic reviews. J Am Med Inform Assoc. 2010;17(4):446–53.
Ji X, Ritter A, Yen P-Y. Using ontology-based semantic similarity to facilitate the article screening process for systematic reviews. J Biomed Inform. 2017;69:33–42.
Cohen AM, et al. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc. 2006;13(2):206–19.
Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Booth A. “Brimful of STARLITE”: toward standards for reporting literature searches. J Med Libr Assoc. 2006;94(4):421.
Hoffmann TC, et al. Better reporting of interventions: template for intervention description and replication (TIDieR) checklist and guide. Bmj. 2014;7:348.
Spiteri L. A simplified model for facet analysis: Ranganathan 101. Can J Inf Libr Sci. 1998;23(1–2):1–30.
Fernandez-Felix BM, et al. CHARMS and PROBAST at your fingertips: a template for data extraction and risk of bias assessment in systematic reviews of predictive models. BMC Med Res Methodol. 2023;23(1):1–8.
Cochran WG. The comparison of percentages in matched samples. Biometrika. 1950;37(3/4):256–66.
Higgins JP, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Gabrys HS, et al. Design and selection of machine learning methods using radiomics and dosiomics for normal tissue complication probability modeling of xerostomia. Front Oncol. 2018;8:35.
Lee TF, et al. Using multivariate regression model with least absolute shrinkage and selection operator (LASSO) to predict the incidence of xerostomia after intensity-modulated radiotherapy for head and neck cancer. Plos One. 2014;9(2):89700.
Lee TF, Liou MH, Huang YJ, Chao PJ, Ting HM, Lee HY, Fang FM. LASSO NTCP predictors for the incidence of xerostomia in patients with head and neck squamous cell carcinoma and nasopharyngeal carcinoma. Sci Rep. 2014;4(1):6217.
van Dijk LV, et al. CT image biomarkers to improve patient-specific prediction of radiation-induced xerostomia and sticky saliva. Radiother Oncol. 2017;122(2):185–91.
Ursino S, et al. Incorporating dose–volume histogram parameters of swallowing organs at risk in a videofluoroscopy-based predictive model of radiation-induced dysphagia after head and neck cancer intensity-modulated radiation therapy. Strahlenther Onkol. 2021;197:209–18.
Dean J, et al. Incorporating spatial dose metrics in machine learning-based normal tissue complication probability (NTCP) models of severe acute dysphagia resulting from head and neck radiotherapy. Clinic Trans Radiat Oncol. 2018;8:27–39.
Dean JA, et al. Normal tissue complication probability (NTCP) modelling using spatial dose metrics and machine learning methods for severe acute oral mucositis resulting from head and neck radiotherapy. Radiother Oncol. 2016;120(1):21–7.
Beetz I, et al. NTCP models for patient-rated xerostomia and sticky saliva after treatment with intensity modulated radiotherapy for head and neck cancer: the role of dosimetric and clinical factors. Radiother Oncol. 2012;105(1):101–6.
Beetz I, et al. Development of NTCP models for head and neck cancer patients treated with three-dimensional conformal radiotherapy for xerostomia and sticky saliva: the role of dosimetric and clinical factors. Radiother Oncol. 2012;105(1):86–93.
Men K, et al. A deep learning model for predicting xerostomia due to radiation therapy for head and neck squamous cell carcinoma in the RTOG 0522 clinical trial. Int J Radiat Oncol Biol Phys. 2019;105(2):440–7.
Rosen BS, et al. Early changes in serial CBCT-measured parotid gland biomarkers predict chronic xerostomia after head and neck radiation therapy. Int J Radiat Oncol Biol Phys. 2018;102(4):1319–29.
Sheikh K, et al. Predicting acute radiation induced xerostomia in head and neck cancer using MR and CT radiomics of parotid and submandibular glands. Radiat Oncol. 2019;14(1):1–11.
Gaito S, et al. Normal tissue complication probability modelling for toxicity prediction and patient selection in proton beam therapy to the central nervous system: a literature review. Clin Oncol. 2022;34(6):e225–37.
Bang C, et al. Artificial intelligence to predict outcomes of head and neck radiotherapy. Clinic Transl Radiat Oncol. 2023;39:100590.
Brown T, et al. Language models are few-shot learners. Adv Neural Inf Process Syst. 2020;33:1877–901.
Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8.
Zheng T, et al. Detection of medical text semantic similarity based on convolutional neural network. BMC Med Inform Decis Mak. 2019;19:1–11.
Ntonti P, et al. A systematic review of reading tests. Int J Ophthalmol. 2023;16(1):121.
Acknowledgements
This study was supported financially, in part, by grants from the National Science and Technology Council (NSTC) of the Executive Yuan of the Republic of China, (110-2221-E-992-005-MY2, 111-2221-E-992-016-MY2). Part of this study has been presented as a thesis in Chinese.
Funding
Grants from the National Science and Technology Council (NSTC) of the Executive Yuan of the Republic of China, (110–2221-E-992–005-MY2, 111–2221-E-992–016-MY2).
Author information
Authors and Affiliations
Contributions
Conceptualization: P-J.C, T-F.L. Data curation: Y-W.H., P-Y. Y., C-H. T., S–H.L., L.C., C-D. T. Methodology: P-J C. J.Y., J-M. W.. Project administration: T-F L. Writing ± original draft: T-F L. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Institutional review board approval was not needed as this study did not involve human participants.
Consent for publication
We hereby confirm that all authors have seen and agree with the contents of the manuscript being submitted. We warrant that the article is the authors' original work, has not received prior publication, and is not under consideration for publication elsewhere. We give our consent for the publication of identifiable details, which can include figure(s) and/or table(s) and the details within it/them, in Radiation Oncology.
Competing interests
All authors have declared that no competing interests exist.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1 Table S1.
Database Retrieval Detail Sheet. Table S2. Optimizer Test Set Performance Comparison Table. Table S3. Screening speed measured in Words Per Minute (WPM). Figure S1. Code for WPM Calculation Algorithm captured from the monitor. Supplementary Figure S2. Bias funnel chart for the a early -effect b late-effect xerostomia
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lee, TF., Hsieh, YW., Yang, PY. et al. Using meta-analysis and CNN-NLP to review and classify the medical literature for normal tissue complication probability in head and neck cancer. Radiat Oncol 19, 5 (2024). https://doi.org/10.1186/s13014-023-02381-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13014-023-02381-7